Liquid biomolecular droplets assembled through phase separation mechanisms
Light-dependent control of phase separation in living cells
Programmable DNA droplets accelerating molecular computation
Living cells are more than a bag of biomolecules. Cells are compartmentalized with various organelles with unique molecular composition and function. For example, the nucleus contains DNA, molecules harboring genetic information, while mitochondria power ATP production through cellular respiration. Similar to the cell itself, these organelles are encapsulated by lipid bilayers. However, cells also contain many examples of organelles without delimiting membrane. These so-called membrane-less organelles include, among many, nucleoli, nuclear speckles, germ granules, and stress granules. From a physical perspective, these structures are highly interesting in that they can maintain specific molecular compositions even in the absence of physical barriers of lipid bilayers. Although membrane-less organelles were discovered more than a century ago, the physical mechanism of driving their assembly was unclear.
Recent studies revealed that membrane-less organelles exhibited liquid-like behaviors1. Their shapes tend to be highly spherical, implying the presence of surface tension. Remarkably, when two membrane-less organelles meet one another, they fuse into a single structure while exhibiting shape relaxation back to the spherical one. The observation of liquid-phase organelles, now termed condensates, within protoplasm led to a hypothesis that cellular content undergoes phase separation to form immiscible coexisting condensates, similar to oil and water. Since 2015, numerous studies have shown biomolecular phase separation acts as a key organizing principle for diverse cellular processes, including gene regulation, stress response, cell signaling, and homeostasis2. In addition, abnormal forms of phase separation are implicated in various disease states, such as cancer and neurodegeneration. Due to its significant relevance to cell physiology and malfunction, huge efforts from the pharmaceutical industry have recently been put forth to develop new types of drugs targeting intracellular phase separation3.
Professor Yongdae Shin, in the Department of Mechanical Engineering and Interdisciplinary Program in Bioengineering, has been developing a series of technologies to control phase separation in living cells. His research findings were published twice in the journal Cell. In 2017, using engineered light-sensitive protein constructs, he demonstrated that intracellular phase separation was spatiotemporally controllable via external light input4. Since then, the technology, termed optoDroplets, has been widely adopted by numerous research groups to study phase behaviors of proteins of interest, screen key protein segments driving phase separation, and build synthetic organelles for metabolic engineering. In 2018, he introduced another light-inducible phase separation system called CasDrop5. This technology was an outcome of integrating optogenetic control of phase separation with CRISPR-Cas9-based gene targeting. Thus, it enabled the controlled nucleation of liquid condensates at specific genomic regions. The underlying idea was similar to artificial rains, where nuclei for water condensation were employed to facilitate heterogeneous nucleation. Using this technology, he showed that physical forces provided by liquid-phase organelles could modulate 3D genome organization.
Prof. Shin and his group have been pursuing research to reveal the physiological functions of intracellular condensates. In particular, a major research thrust of the group is to understand the role of biomolecular phase separation on gene expression. Notably, the aberrant formation of condensates driven by oncofusion proteins is frequently observed in numerous cancers. Funded by Samsung Science and Technology Foundation, his group combines multiple cutting-edge experimental technologies, such as optogenetics and spatial multi-omics, to dissect the mechanism of condensate functions. Another focus of his research is to probe the relations between the physical properties and functions of condensates. They have been using quantitative phase imaging to measure the density of liquid-phase organelles within living cells. Tuning the physical properties of condensates can provide an effective way of modulating the physiological consequences of condensate-mediated cellular processes.
Prof. Shin has also been engineering synthetic liquid-phase organelles to harness emergent properties of biomolecular phase separation. In a project funded by the National Science Challenge Initiatives Program, his research group sought to achieve synthetic condensates with programmable organization and functions. To accomplish this, they used DNA as a building block. Taking advantage of the unique capability of DNA in defining specificity and strength of intermolecular interactions, the group successfully implemented DNA-based synthetic droplets enriched in a set of DNA molecules with specific sequences6. They then coupled DNA-based molecular computation to the synthetic droplets, demonstrating that molecular computation reactions were drastically accelerated. This work, published in the journal Science Advances, highlights the emergent functions that originated from collective interactions between phase-separating components.
Over the past decade, the study of biomolecular phase separation has attracted huge attention in the biomedical research field. It is an exciting topic where multiple disciplines can collectively make an enormous impact. Prof. Shin envisions that as basic sciences further reveal fundamental principles of biomolecular phase separation, an era of organelle engineering will follow to fully harness the emergent behaviors of biomolecules for improving human health and wellness.
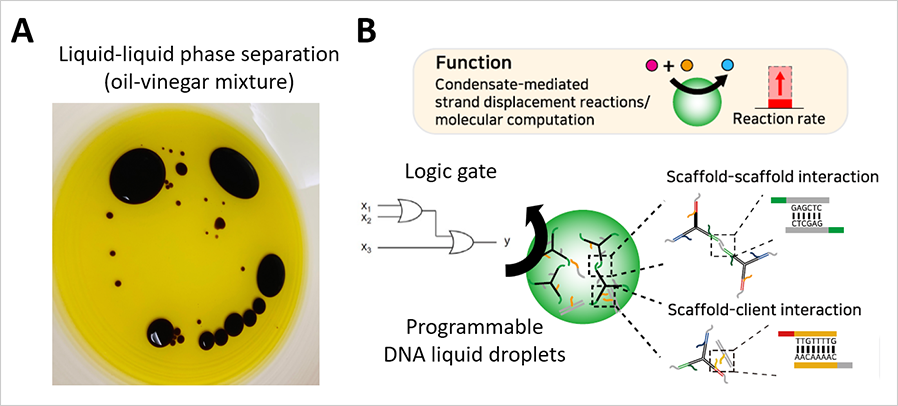
Figure
(A) Phase separation in our daily life.
(B) Programmable DNA liquid droplets boosting up the speed
of molecular computation
References
1. C.P. Brangwynne et al., Science 2009, 324, 1729-1732.
2. Y. Shin et al., Science 2017, 357, eaaf4382.
3. D.M. Mitrea et al., Nat Rev Drug Discov 2022, 1-22.
4. Y. Shin et al., Cell 2017, 168, 159-171.
5. Y. Shin et al., Cell 2018, 175, 1481-1491.
6. S. Do et al., Science Advances 2022, 8, eabj1771.
1. C.P. Brangwynne et al., Science 2009, 324, 1729-1732.
2. Y. Shin et al., Science 2017, 357, eaaf4382.
3. D.M. Mitrea et al., Nat Rev Drug Discov 2022, 1-22.
4. Y. Shin et al., Cell 2017, 168, 159-171.
5. Y. Shin et al., Cell 2018, 175, 1481-1491.
6. S. Do et al., Science Advances 2022, 8, eabj1771.


